如何可视化家系间的亲缘关系
- 运行前清除掉原来的一些对象变量,然后加载包。
- optiSel(处理系谱数据、计算个体间的共亲系数)、dplyr(清洗数据)、data.table(数据框存为该格式,加快清洗速度)和reshape2(清洗数据)。
rm(list=ls())
knitr::opts_chunk$set(echo = TRUE,cache = FALSE,warning=FALSE,message=FALSE,dpi = 600,fig.width = 8, out.width = "70%", fig.align = "center")
require(optiSel) #系谱处理和计算共亲系数
require(dplyr) #数据清洗
require(data.table) #加快计算速度
require(reshape2) #数据变形
require(magrittr) #管道符
require(circlize) #加载circlize包画弦线图
1.circlize简介
circlize (https://cran.r-project.org/web/packages/circlize/) 是一个华人(Zuguang Gu)开发的包,是对用于基因组数据进行展示的circos (http://circos.ca/) 软件功能的一个R实现包。
circlize的功能(http://zuguang.de/circlize_book/book/) 非常多,这里我们主要是利用它解析家系间的亲缘关系。
2. circlize使用
2.1 数据读取
演示数据主要来自于一种虾的系谱。首先利用R自带的read.table函数读取系谱信息
test.ped <- read.table(file="Data/Input/TestPed.csv",header = TRUE,sep=",",stringsAsFactors = TRUE)
tail(test.ped)
## AnimalID SireID DamID FamilyID SexID Year Breed
## 5017 G010D804 G900B326 G900E185 16F981 2 2016 unknown
## 5018 G010D920 G900B326 G900E185 16F981 2 2016 unknown
## 5019 G010D929 G900B326 G900E185 16F981 2 2016 unknown
## 5020 G010D931 G900B326 G900E185 16F981 1 2016 unknown
## 5021 G010D964 G900B326 G900E185 16F981 2 2016 unknown
## 5022 G0100033 G900B326 G900E185 16F981 2 2016 unknown
2.2 系谱分析和亲缘系数计算
- 为了加快计算速度,将数据框转换为data.table格式
- 利用optiSel包中的prePed函数对系谱进行整理,主要目的是对系谱按照祖先在前,个体在后的顺序进行排序;
- 利用optiSel包中的pedIBD函数计算2016任意两个体间的共亲系数;
#prePed函数来自optiSel
test.ped.pre <- prePed(test.ped)
#筛选2016年个体
keep <- test.ped.pre$Indiv[test.ped.pre$Year == 2016]
#在计算个体间的共亲系数前,每个家系只入选一个个体做代表,整体计算量就下来了。
unique.indiv <- distinct(test.ped.pre[keep,],Sire,Dam,Sex, .keep_all = TRUE)$Indiv
#计算2016年家系个体间的共亲系数
keep.kin <- pedIBD(test.ped.pre, keep.only = unique.indiv)
#keep.kin是一个对称矩阵
class(keep.kin)
## [1] "matrix"
- 数据整理为3列形式
#转为data.table格式
keep.kin.data.table <-
as.data.table(keep.kin, keep.rownames = TRUE)
#利用reshape2包中的melt函数将矩阵转为3列形式
keep.kin.three.column <-
melt(
keep.kin.data.table,
id.vars = c("rn"),
measure.vars = colnames(keep.kin.data.table)[-1]
)
colnames(keep.kin.three.column) <- c("ID1", "ID2", "cc")
head(keep.kin.three.column)
## ID1 ID2 cc
## 1: G010A022 G010A022 0.52929688
## 2: G010A002 G010A022 0.07714844
## 3: G010A528 G010A022 0.07714844
## 4: G010A204 G010A022 0.11425781
## 5: G010A050 G010A022 0.09277344
## 6: G010A348 G010A022 0.00000000
max(keep.kin.three.column$cc)
## [1] 0.5332031
keep.kin.three.column$ID2 <-
as.character(keep.kin.three.column$ID2)
#利用管道符号 %>% 简化代码书写和计算流程
#找出每个个体所在的家系
keep.kin.three.column.family <- keep.kin.three.column %>%
left_join(., test.ped.pre[, c("Indiv", "FamilyID")], by = c("ID1" = "Indiv")) %>%
left_join(., test.ped.pre[, c("Indiv", "FamilyID")], by = c("ID2" = "Indiv"))
#去除重复的家系组合,理论上不会再有重复
keep.kin.three.column.family.unique <-
keep.kin.three.column.family[, c("FamilyID.x", "FamilyID.y", "cc")] %>%
distinct(.,
FamilyID.x,
FamilyID.y,
cc,
.keep_all = TRUE)
head(keep.kin.three.column.family)
## ID1 ID2 cc FamilyID.x FamilyID.y
## 1 G010A022 G010A022 0.52929688 16F1191 16F1191
## 2 G010A002 G010A022 0.07714844 16F1201 16F1191
## 3 G010A528 G010A022 0.07714844 16F1202 16F1191
## 4 G010A204 G010A022 0.11425781 16F1211 16F1191
## 5 G010A050 G010A022 0.09277344 16F1241 16F1191
## 6 G010A348 G010A022 0.00000000 16F011 16F1191
2.3 用circlize画圈图
2.3.1 选取10个家系,绘制它们间的亲缘系数
ten.families <- unique(keep.kin.three.column.family.unique$FamilyID.x)[1:10]
keep.kin.three.column.family.ten <- keep.kin.three.column.family.unique %>%
filter(., (FamilyID.x %in% ten.families) & (FamilyID.y %in% ten.families))
chordDiagram(keep.kin.three.column.family.ten)
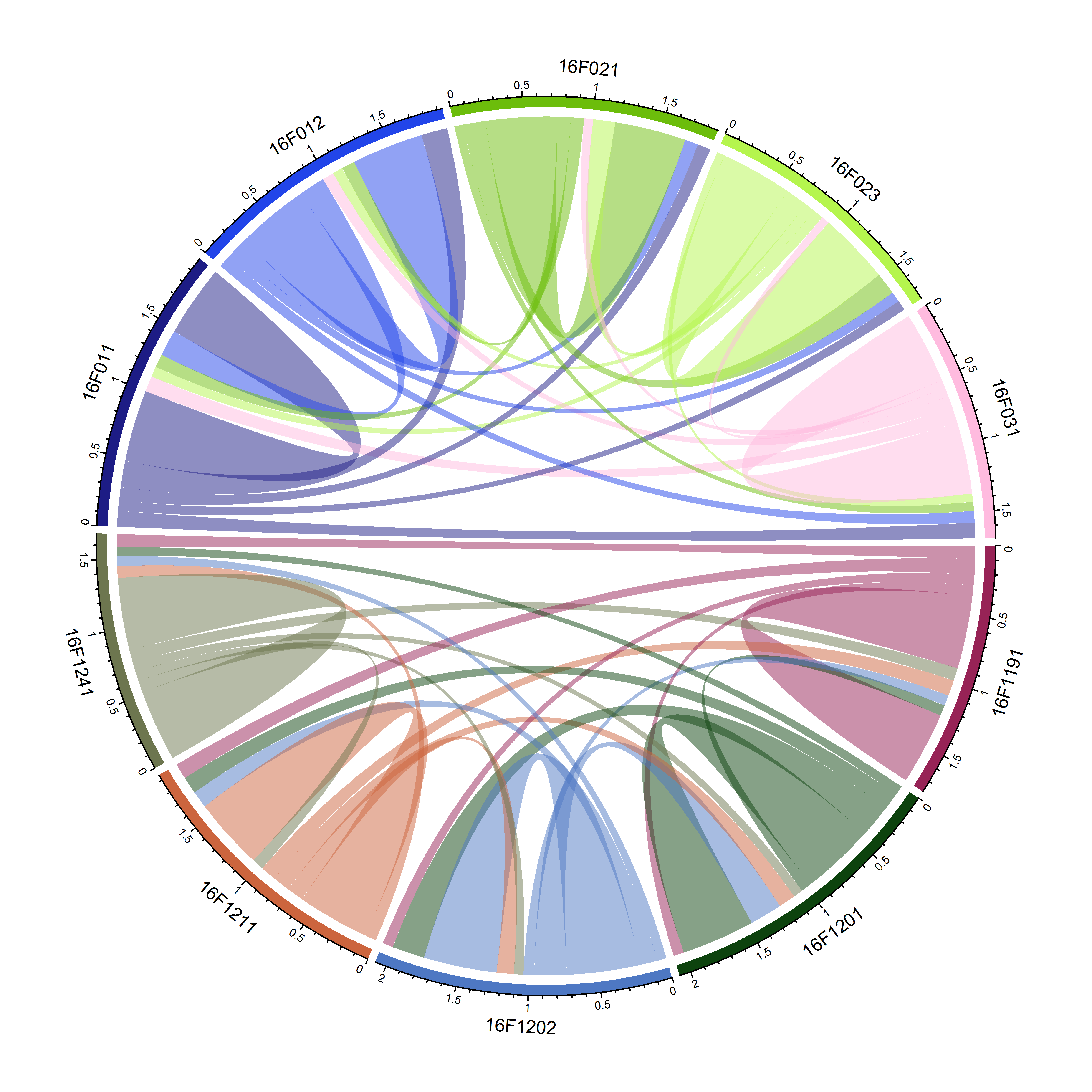 图1 10个家系间的共亲系数弦状图
图1 10个家系间的共亲系数弦状图
circos.clear()
sort(unique(keep.kin.three.column.family.ten$cc))
## [1] 0.00000000 0.05923021 0.06555462 0.06933594 0.07493985 0.07714844
## [7] 0.08462715 0.08496094 0.09277344 0.09562135 0.11425781 0.11494756
## [13] 0.12402344 0.16125262 0.19129539 0.23632812 0.51935029 0.52068853
## [19] 0.52125573 0.52234769 0.52343750 0.52929688 0.53320312
2.3.2 清除自我连接后的家系亲缘关系弦状图
- 从上图中可以看出,每个家系自我连接。在不考虑个体自身近交系数的情况下,主要是指个体自身的共亲系数0.5。 下边的代码将会清除家系间的自我连接。
keep.kin.three.column.family.ten.between <- keep.kin.three.column.family.ten %>%
filter(.,!(FamilyID.x==FamilyID.y))
sort(unique(keep.kin.three.column.family.ten.between$cc))
## [1] 0.00000000 0.05923021 0.06555462 0.06933594 0.07493985 0.07714844
## [7] 0.08462715 0.08496094 0.09277344 0.09562135 0.11425781 0.11494756
## [13] 0.12402344 0.16125262 0.19129539 0.23632812
chordDiagram(keep.kin.three.column.family.ten.between)
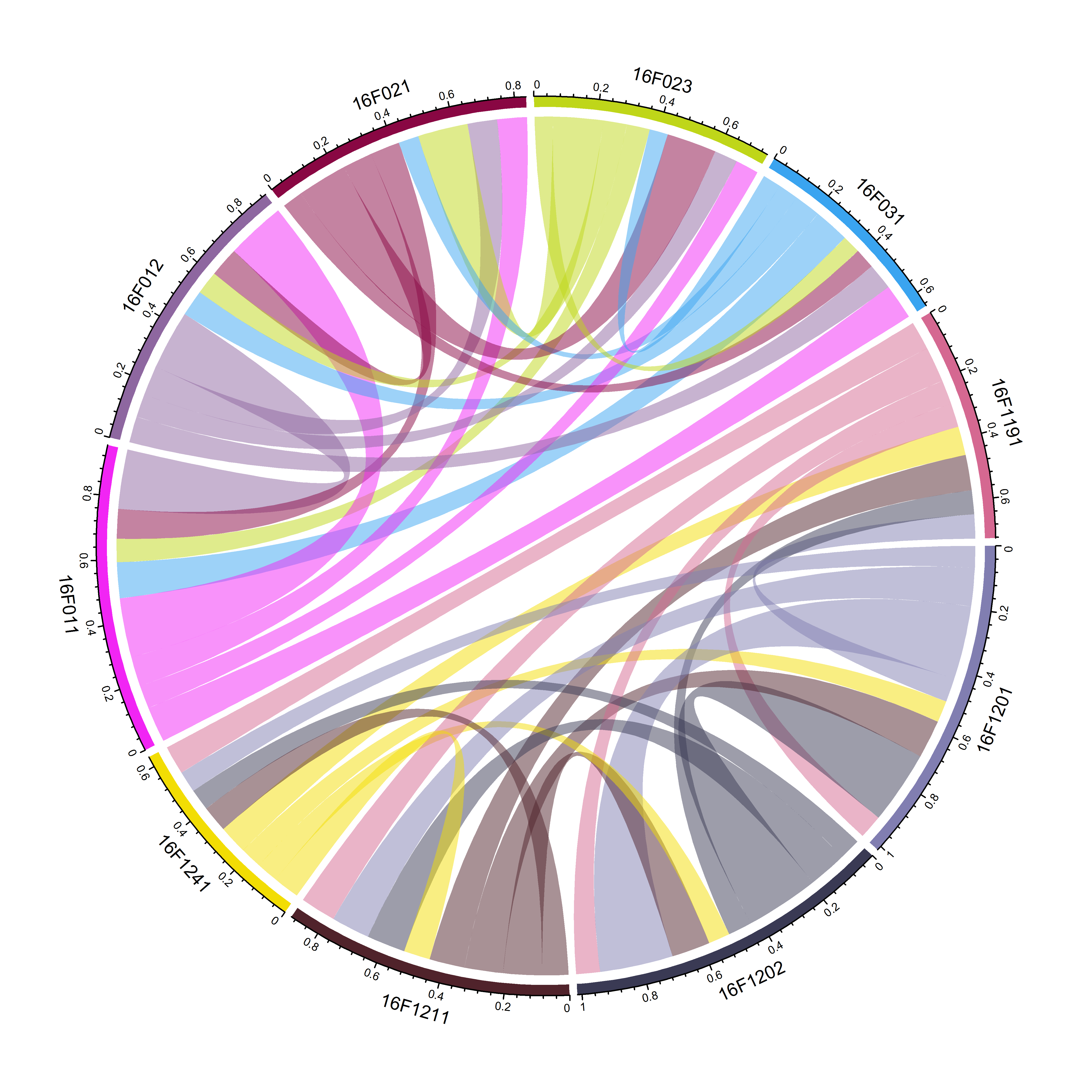 图2 10个家系间的共亲系数弦状图(排除家系内的共亲系数)
图2 10个家系间的共亲系数弦状图(排除家系内的共亲系数)
circos.clear()
2.3.3 设定区块和连接线的颜色
circlize生成的区块和连接线颜色,都是随机的。如果想要使用自己设定的颜色,需要设定grid.col参数,生成颜色可以有多种颜色函数,这里用的是rainbow(),来自R自带的包grDevices
chordDiagram(keep.kin.three.column.family.ten.between,grid.col = rainbow(10))
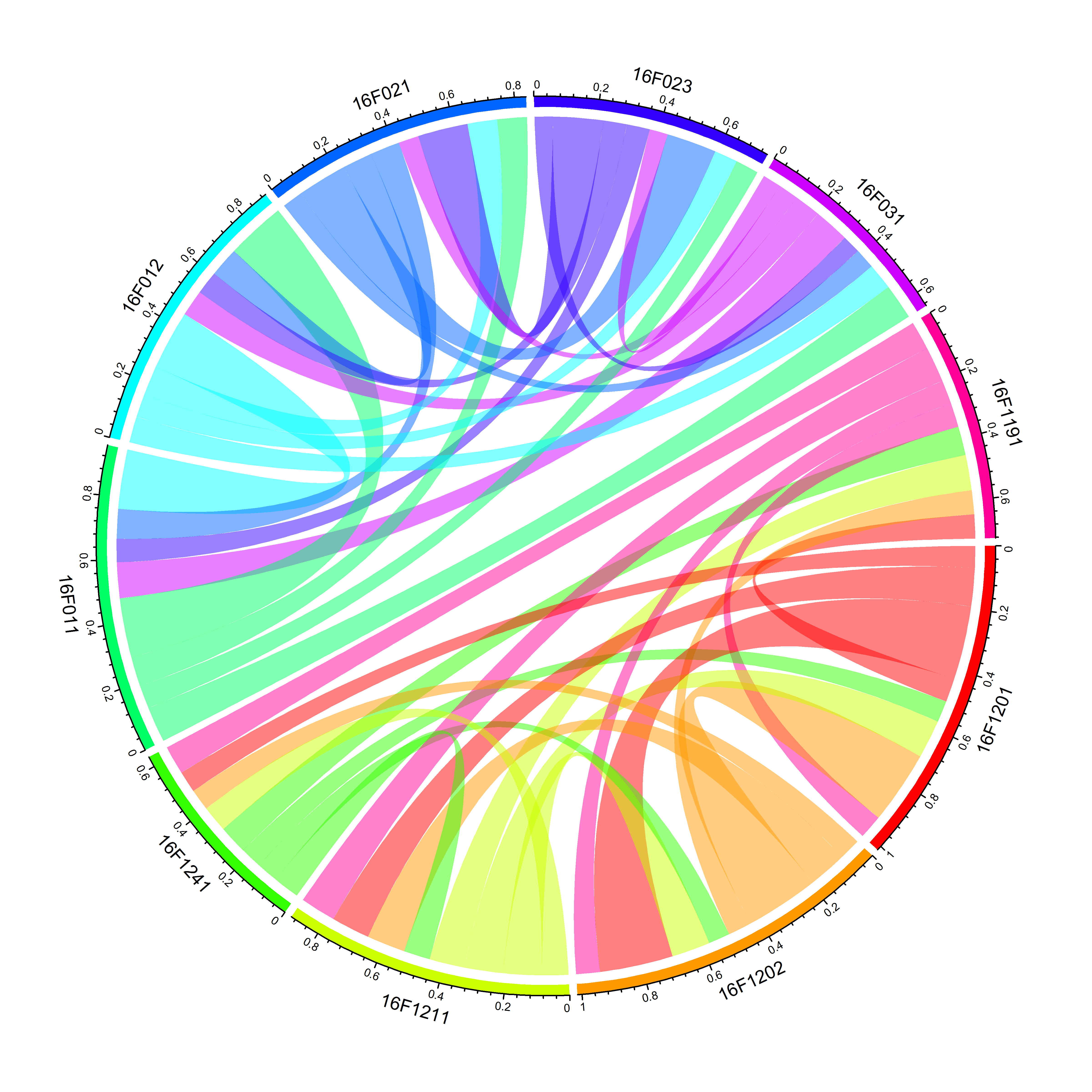 图3 设定区块和连接线的颜色(grid.col参数)
图3 设定区块和连接线的颜色(grid.col参数)
circos.clear()
2.3.4 一种简单的作图形式。其实前边的对称矩阵keep.kin可以不用再去变为3列矩阵,也不需要去掉重复等,可以直接用对称矩阵作图。
symmetric=TRUE表示矩阵是对称的,只使用下三角矩阵。
keep.kin.ten.family.sym <- keep.kin[ten.families,ten.families]
chordDiagram(keep.kin.ten.family.sym,grid.col = rainbow(10),symmetric = TRUE)
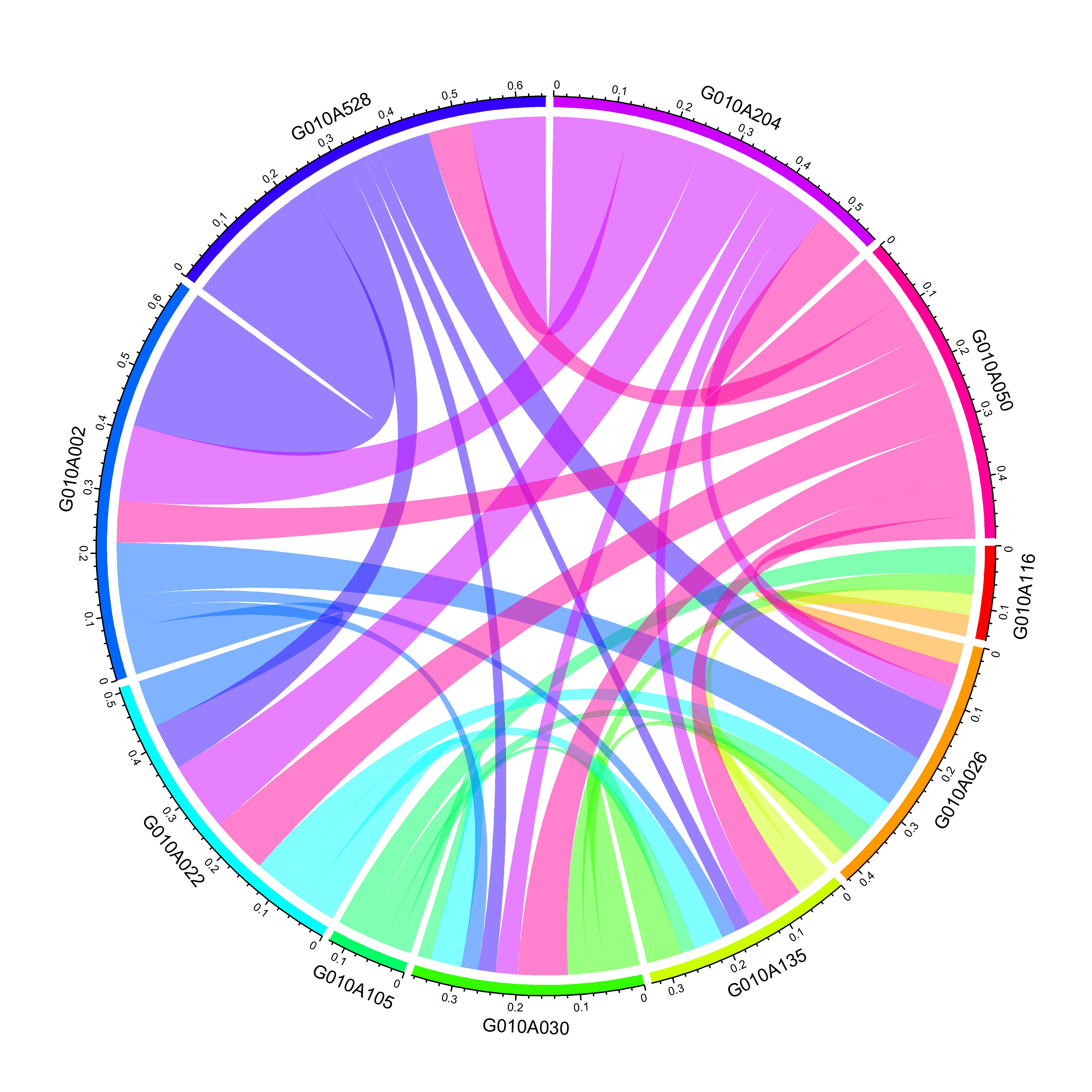 图4 画出家系间的亲缘关系(排除家系自身亲缘系数)
图4 画出家系间的亲缘关系(排除家系自身亲缘系数)
chordDiagram(keep.kin.ten.family.sym,grid.col = rainbow(10),symmetric = FALSE)
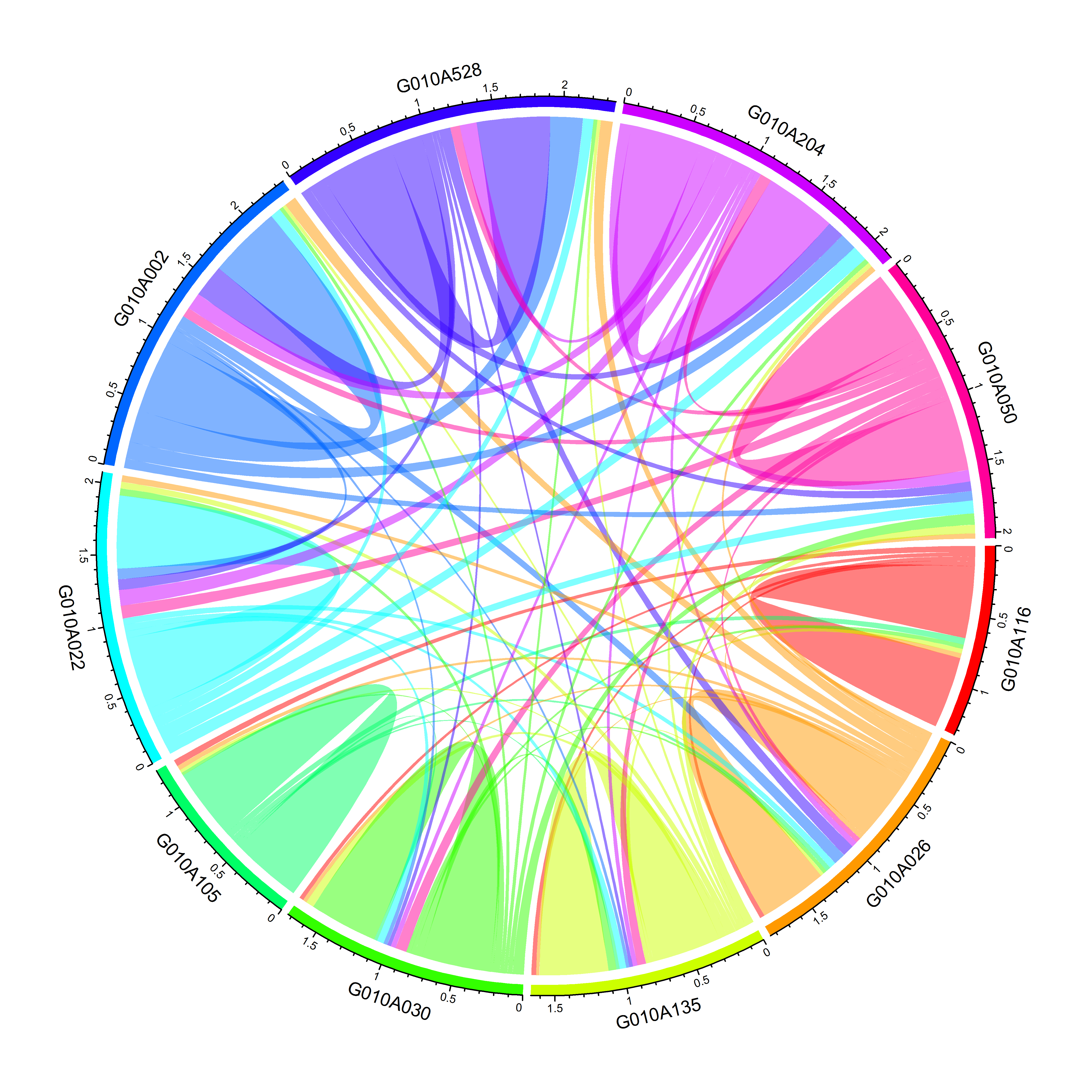 图5 画出家系间的亲缘关系(包括家系自身亲缘系数)
图5 画出家系间的亲缘关系(包括家系自身亲缘系数)
- 画出全部家系间的弦状图(这个图非常耗费时间,在chunk中设置eval=FALSE屏蔽了,只展示代码)
keep.kin.three.column.family.unique.no.within <- keep.kin.three.column.family.unique %>% filter(., !(FamilyID.x == FamilyID.y)) chordDiagram(keep.kin.three.column.family.unique.no.within) circos.clear()
- 上一篇 my first blog
- 下一篇 Rcpp读书笔记:第一章Rcpp简介